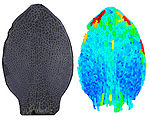
Shaping Diversity
How do small groups of cells in microscopic buds turn themselves into the diverse flower and leaf shapes we see around us? To answer this question we need to know how genes and growth interact to create tissue shapes during development, and how this process varies to produce such a remarkable range of forms. We use a highly integrative approach that combines molecular, genetic, imaging, population, ecological and computational approaches to address this problem, applying them to model systems such as Arabidopsis and Antirrhinum, as well as the model carnivorous plant Utricularia.