Microtubule dynamics during growth and division
Jordi Chan in collaboration with Richard Kennaway

Microtubules are highly dynamic subcellular filaments that adopt different patterns of alignment during growth, division and differentiation. During growth, microtubules are found in close association with the plasma-membrane where they are needed to guide the movement of cellulose synthase to generate appropriate wall architectures. Like animal cells, microtubules are required for chromosome segregation during cell division via the spindle apparatus. However, unlike animal cells, plant microtubules bunch up at the onset of mitosis to form a cortical ring, known as the pre-prophase band (PPB), which is involved in determining the future plane of cell division. Since plant cells within tissues are immobilised by their own walls and can only respond to cellular and environmental cues by altering their directions of growth and division planes, microtubules are indispensible elements of plant morphogenesis.
How microtubules change alignment and construct different arrays during growth and cell division is unknown but is likely to reside within fundamental properties of the microtubules themselves, such as the dynamic behaviour of their filamentous ends, regulation of their sites and angles of nucleation (birth) and outcomes of encounters between themselves and the geometry of the cell. It is interesting to think that these simple subcellular behaviours or rules concerning microtubule dynamics may not only shape the cell but also be amplified during morphogenesis from cell-to-cell to shape the entire plant.
The aim of my work is to reconstitute microtubule dynamics and cortical array organisation in-silico to understand and test mechanisms behind microtubule reorientation and division plane alignment (PPB formation) in-planta. This will involve time-lapse microscopy of growing and dividing leaf cells expressing appropriate fluorescent reporter genes, the development of software to quantify microtubule dynamics at both subcellular and tissue levels, and computer modelling. Models will be tested using cytoskeletal-specific drugs and mutants harbouring defective cytoskeletal genes. I will also apply these methods to other systems such as Utricularia.
Genetic analysis of plant growth and development
Karen Lee in collaboration with Rob Bellow and Richard Smith

Leaves come in many shapes and sizes, from flat leaves with a simple outline or serrated edges, to the cup shaped traps of carnivorous plants.
My research focuses on understanding how genetic interactions and differential growth act to shape asymmetries in the simple leaves of Arabidopsis and generate complex 3D shapes such as those of carnivorous plant traps.
Using a combination of genetics, confocal microscopy, and Optical Projection Tomography my work aims to find whether common rules might underly the diversity of leaf shape seen in nature using Arabidopsis and bladderwort traps of Utricularia gibba as a model system.
Modelling Growth and Development

I am developing finite element methods for modelling the growth and development of tissues such as leaves and petals. The interactive software tool I am developing, called GFtbox, is available for download.
Evolutionary dynamics underlying species diversification
Matthew Couchman in collaboration with Lucy Copsey, Des Bradley and Daniel Richardson

We are part of a collaboration investigating factors affecting gene flow across populations. The project focuses on flower colour in two distinct subspecies of Antirrhinum. Within the Spanish Pyrenees there are a number of hybrid zones that provide ideal environments for analysing how genes influence flower colour. For each individual within our chosen hybrid zone we are annually recording their GPS location and colour scores as well as taking samples for genotyping and other molecular analysis.
My role within the project is to develop a relational database to capture these and other collaborator outputs as well as a website to act as a gateway to this database. The website will include visual tools such as a configurable map of recorded GPS positions and genetic and physical maps.
Natural variation of flower colour
Desmond Bradley in collaboration with Annabel Whibley, David Field, Christophe Andalo and Monique Burrus

Two subspecies of Antirrhinum (magenta and yellow), which generally occur in genetic isolation in the Pyrenees, have hybridized in nature to create a hybrid zone. Due to hybridization the Antirrhinum flowers display an array of parental as well as mixed flower colour phenotypes which are the result of genetic variation at three key loci ROSEA, ELUTA and SULFUREA. The two first genes are involved in the control of the magenta anthocyanin pigmentation while SULF is a repressor of aurone (yellow) pigmentation. As the HZ gives a great playground to study evolution in action, the goal of this research is to follow the genetic flow of these flower colour loci within the hybrid population as well as the fitness of each phenotype/genotype throughout several years. I also use similar approaches to analyse gene expression regulation in Utricularia.
Evolution of novel phenotypes through regulatory small RNAs
Daniel Richardson in collaboration with Des Bradley, Lucy Copsey, Matt Couchman, Annabel Whibley, Tamas Dalmay, Simon Moxon, and Leighton Folkes

Neofunctionalisation (NF) is the process by which, following a duplication event, a given gene gains a new function. This process is typically split into two classes; coding NF refers to situations where the new function arises within the coding sequence of the duplicated gene, and regulatory NF refers to instances where the new function manifests due to changes in the expression pattern of the gene.
Study of a natural hybrid zone of two subspecies of the garden snapdragon Antirrhinum majus has revealed that, within a subset of the population, positive selection is acting on a locus containing an inverted duplication of FLA, a gene involved in the synthesis of the yellow pigment aurone. This locus, SULF, appears to have arisen relatively recently in evolutionary time from multiple duplications of FLA, and acts to control patterns of yellow pigmentation on flowers through the generation of FLA-inhibiting regulatory small RNAs (sRNAs). SULF represents an interesting case study for a novel type of NF; although the duplication events have resulted in a change in the coding sequence of the FLA paralogue, the end result is a change in the expression pattern of the non-neofunctionalised FLA gene. It is presently unclear to what extent this mechanism, dubbed sRNA-mediated neofunctionalisation (SNF), is important in a wider evolutionary context.
My PhD project will utilise genomics and phylogenetics to investigate the involvement of SNF in the evolution of novel phenotypes in wild relatives of A. majus. SNF candidate loci will be identified through analysis of genome and transcriptome data from a range of morphologically diverse Antirrhinum species. The evolutionary history of validated candidates will be examined using phylogenetics. Taken together, these analyses should inform about the extent to which SNF has been involved in the generation of variation in wild snapdragons, how SNF regulatory elements coevolve with their targets, and how general the mechanism is for different traits.

My work is funded by a BBSRC Doctoral Training Partnership studentship.
Polarity of Stomatal Lineage Proteins in Arabidopsis Cotyledon Development
William Bezodis in collaboration with Jordi Chan and John Fozard

Polarity is known to be crucial to plant organ development including flowers and leaves. Cotyledons or seed leaves develop somewhat differently to most leaves and are present in the seed even before germination. Asymmetric cell divisions are required for proper development of the stomatal lineage which gives rise to many of the cells in leaves and cotyledons, polarly localised proteins are important for these asymmetric divisions and can be used to infer polarity across the tissue since the polar localisation must be in response to some sort of signal.
My project looks makes use of these stomatal lineage proteins that have been fused to florescent tags allowing their location to be visualised via confocal microscopy. This can then be used to try and work out the direction and nature of polarity across the cotyledon and how it may or may not be impacted by things like developmental stage, size, and mechanical forces. I am also interested in trying to apply these tools that reveal intrinsic polarity to other parts of plant development where asymmetric divisions are crucial to correct cell identity, as in the stomatal lineage, and will be working on this in collaboration with the Dickinson Lab at the University of Oxford in pollen development.
How do plants adapt to different ecological environments?
Tingting Li in collaboration with Desmond Bradley and Daniel Richardson

Plants evolve different traits for better survival. The Coen lab has been studying various traits of snapdragons (Antirrhinum) such as flower colour, leaf shape and growth habit. These traits differ between different Antirrhinum species and our hypothesis is that these differences have been selected for survival. Our lab has found that some traits show evidence of selection at the genome level, using wild populations of snapdragons.
I am using bioinformation, genetics and molecular biology to explore the adaptive story of snapdragon populations and reveal how different traits are used in different environments.
Rotation of growth through polarity
Zhengting (Tina) Yang in collaboration with Jordi Chan
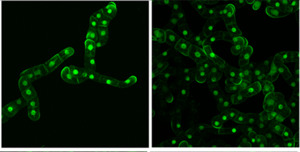
Another hidden feature of cells that controls growth and which may influence the system described above is polarity. Cells can appear uniform, but their contents tend to be organised in an asymmetric manner. A key feature of cell morphogenesis and organismal development is cell polarity. Stomatal asymmetric cell division is regulated by BASL. Tina is interested in understanding the fundamental principles behind how this system is controlled.
Polarity arises from the accumulation of certain molecules at one end of the cell and not the other. These polarity molecules can be coordinated so, that in a tissue, all cells have the polarity molecules at the same end. This collective behaviour is called tissue polarity, and this may be used to orient growth of the tissue.
Tina is working on the hypothesis that polarity is important for steering growth, through regulating interactions with microtubules and cellulose synthases.